In recent years, microbiology has undergone a technological revolution. No longer limited to petri dishes and gram stains, today’s microbial science is powered by artificial intelligence, high-throughput genomics, and robotic automation. This new era often referred to as next-generation microbiology is unlocking deeper insights into the microbial world, accelerating diagnostics, and paving the way for precision applications in healthcare, agriculture, and environmental science
A roadmap for next-generation functional microbiology
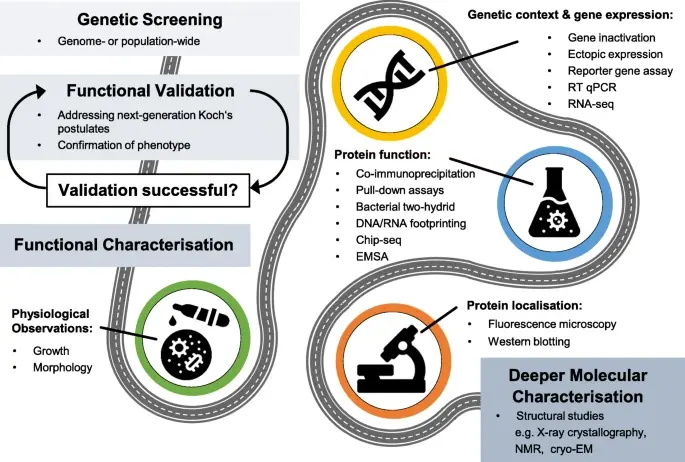
Identifying and then proving that a genetic variant causes a phenotypic change is a considerable step towards understanding bacterial genomics. However, determining the precise role of the gene and how the encoded protein may function requires further study. This can be challenging, particularly for researchers with no background in molecular microbiology, even when the genetic methods or biochemical assays require fairly basic laboratory equipment. Therefore, just as laboratory microbiologists are encouraged to embrace contemporary genomic approaches, so bioinformaticians might consider the central role of functional microbiology in various ways
AI in Microbiology: Smart Insights from Big Data
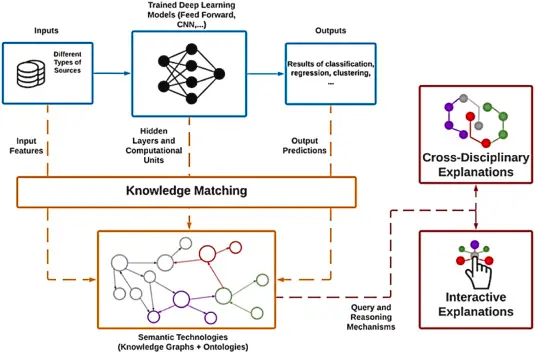
Artificial intelligence is transforming how microbiologists interpret complex datasets. AI algorithms can rapidly analyze large volumes of genomic and metagenomic data, identifying microbial species, predicting antimicrobial resistance, and even uncovering previously unknown pathogens.
Use case examples:
- Clinical diagnostics: AI-powered tools like DeepMicro or PathoScope can detect pathogens directly from sequencing reads, cutting diagnosis time from days to hours.
- Environmental monitoring: Machine learning models help predict microbial behavior in changing ecosystems, assisting in climate and pollution studies.
- Antibiotic discovery: Deep learning is used to screen compound libraries for novel antimicrobials based on microbial genome interactions.
Genomics and Metagenomics: Seeing the Unseen
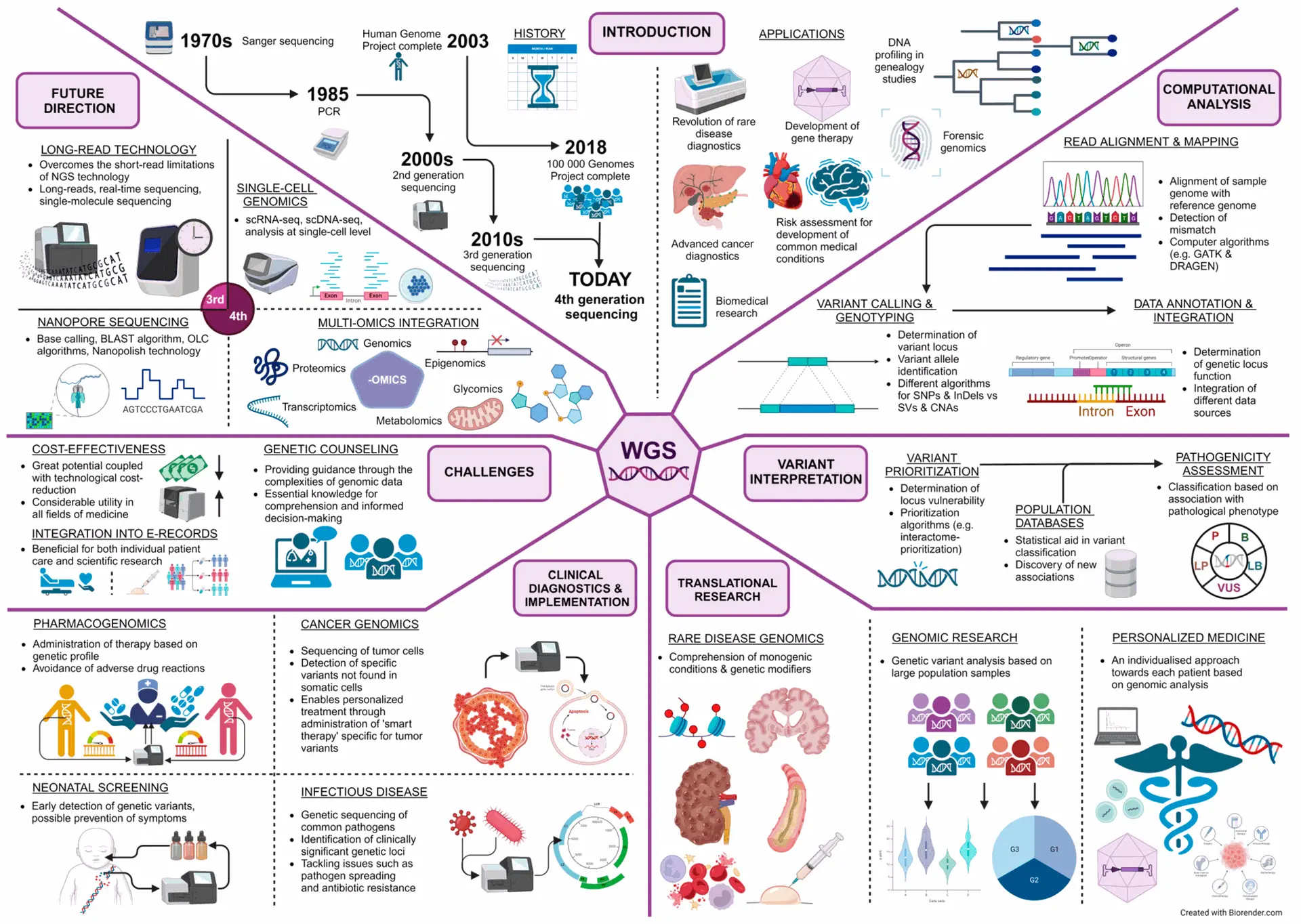
Whole Genome Sequencing (WGS) is revolutionizing microbiology by offering unprecedented resolution in analyzing both human and microbial genomes. It enables precise identification of pathogens, tracking of antimicrobial resistance, and deeper insight into microbial evolution and gene expression.
Beyond microbes, WGS can detect single nucleotide polymorphisms (SNPs) in host DNA variations associated with disease susceptibility, immune response, and treatment outcomes. This dual application makes WGS a powerful tool for studying host–pathogen interactions, understanding microbiome dynamics, and designing personalized antimicrobial therapies.
As WGS becomes more cost-effective and accessible, it’s accelerating diagnostics in clinical microbiology reducing the time to identify rare infections and uncovering genetic variants that impact disease severity. Though challenges remain in interpreting complex data like variants of uncertain significance (VUS), collaborative databases and AI tools are rapidly advancing our ability to turn sequencing data into actionable insights.
In the era of next-generation microbiology, WGS bridges the gap between genomics and infectious disease control empowering precision medicine, outbreak surveillance, and microbiome-based therapies.